A Helpful Handbook to the Human Genome
Each cell in our bodies contains a sequence of DNA that is uniquely ours. This sequence is made up of four kinds of chemicals, known by the letters: A, C, G, and T.
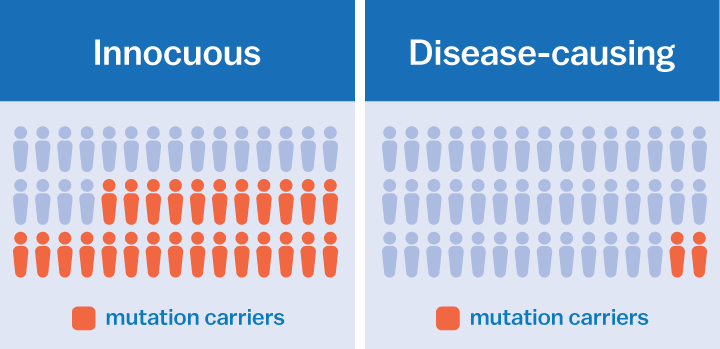
Our genome—the complete set of our genes—comprises three billion of these letters. Thinking of our genome as this enormous set of letters is one way of understanding genetic mutations. Basically, mutations are misspellings. Just as misspelled words happen when a letter is swapped out for another, a genetic mutation occurs when one or more of the four letters aren’t where they should be.

Sometimes these misspellings are innocuous; they don’t make a difference in how our genes behave. But sometimes they are devastating. For instance, a single misspelling can cause diseases like cystic fibrosis or Duchenne muscular dystrophy.

Scientists grasped the potential power of a single genomic spelling mistake decades ago, but they still don’t have an easy and consistently accurate way of answering a related question of obvious importance: how do we know which mutations cause disease—and which ones are harmless?
An international effort—led by Daniel MacArthur, an institute member at the Broad Institute of MIT and Harvard—is hoping to change that. Two years ago, MacArthur and his collaborators launched the Exome Aggregation Consortium (ExAC), a group with the express purpose of collecting sequenced exomes—the parts of the genome made up of genes that encode proteins.
By comparing tens of thousands of these exome sequences—from both patients and healthy participants—the ExAC team has built the beginnings of a revolutionizing reference database: a spelling guide that helps scientists understand which letters are supposed to be where in healthy patients, and which misspellings are likely to represent a disease-causing mutation.
For example, using the data ExAC has gathered, scientists have demonstrated that mutations previously associated with multiple sclerosis or prion disease, a fatal brain disorder, were actually not harmful after all. These mutations occurred too often in healthy populations to be the cause of these particular disorders.
Recently, MacArthur’s team bolstered this already powerful resource by adding entire genomes to the database. The new resource, the Genome Aggregation Database, or gnomAD, contains sequences from more than 140,000 people—and continues to grow. And the more it grows, the more it has the potential to become an essential handbook for gene-based precision medicine.
“This project would have been impossible without the willingness of over a hundred research groups to contribute their data, and the Broad Institute's support and computational power,” MacArthur said. “The resulting resource has impacted the lives of tens of thousands of rare disease patients, demonstrating the incredible power of data sharing.”
